Reference sequences/databases guide
Kathleen Zeglinski
2022-11-15
Introduction
This guide will explain how to prepare reference sequences and databases for NAb-seq (v0.2). If you are using mouse or rat then there is no need to follow this guide, as the references are already included within NAb-seq.
Preparing the all-antibody reference file
Visit the IMGT/GENE-DB website
Select your species, and select the ‘IG’ option under ‘Molecular component’. Then, click submit
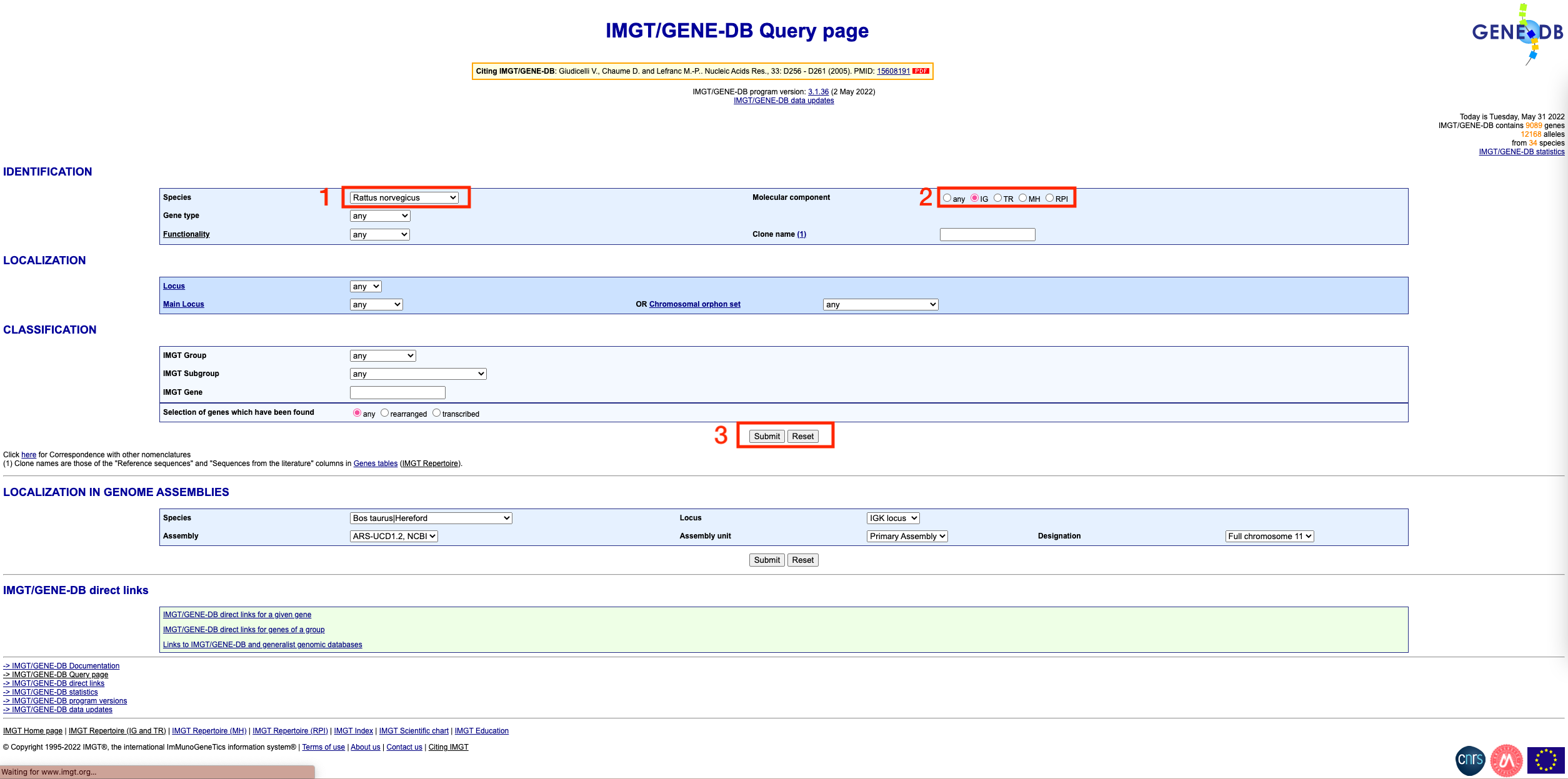
You should see a page that looks like this:
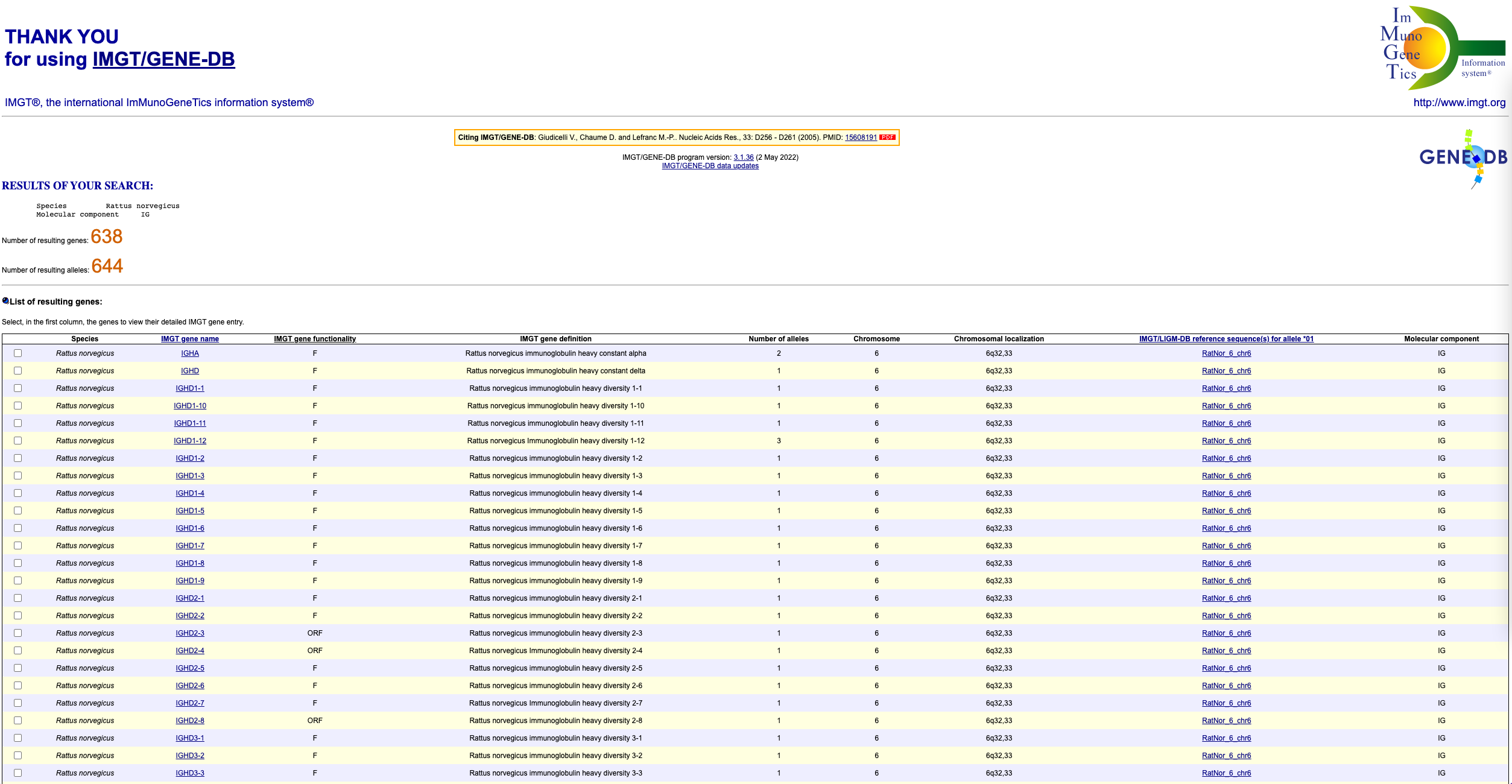
To get the sequences in fasta format, scroll down to the bottom of the page and click on the ‘select all genes’ tickbox. Then, choose ‘F+ORF+all P nucleotide sequences’ under the ‘IMGT/GENE-DB allele reference sequences in FASTA format’ section. Finally, click submit.
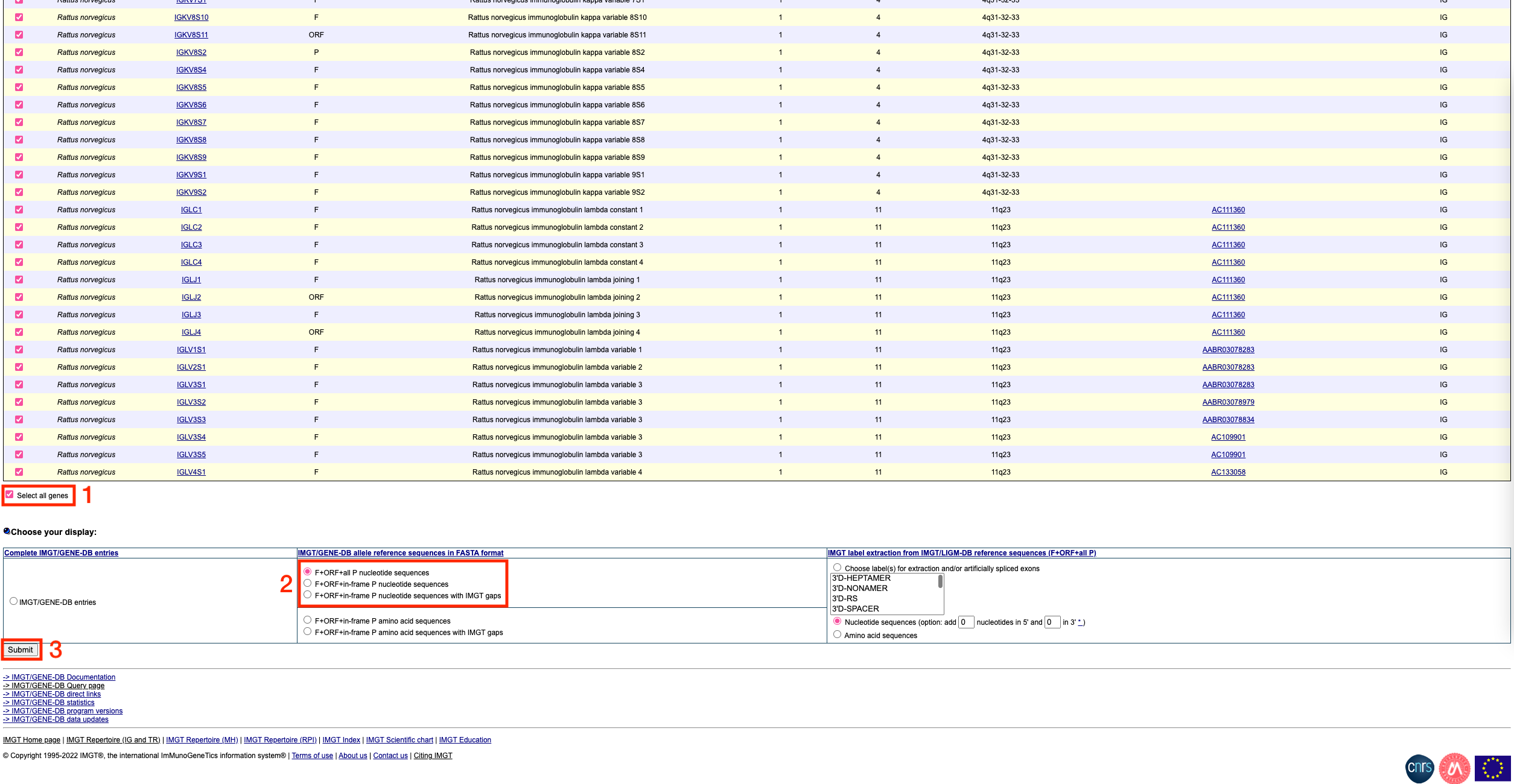
You should see a page that looks like this:
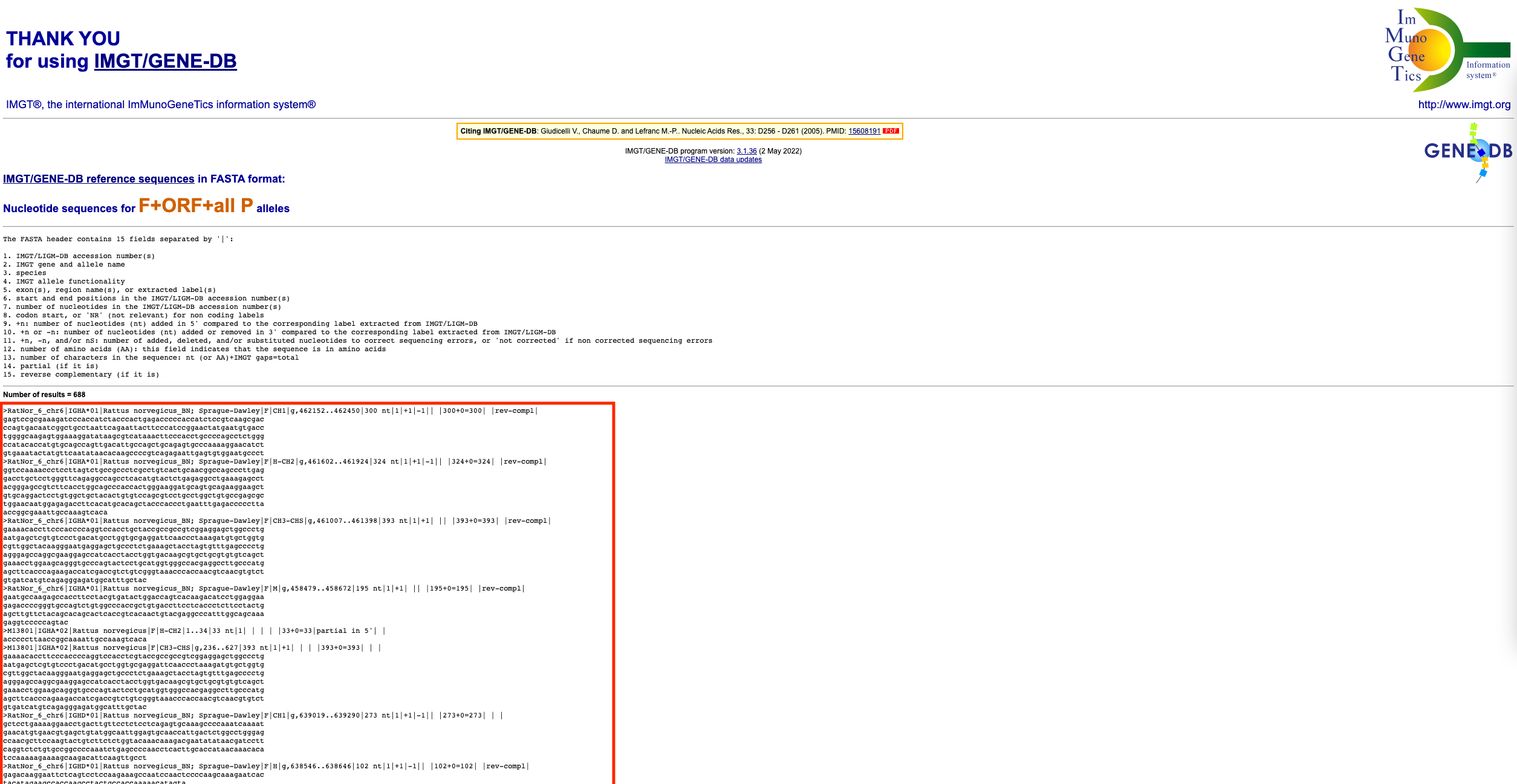
Copy the fasta format sequences (everything below the ‘Number of results’ line, as indicated by the red box above. Paste these sequences into a text editor (e.g. TextEdit on Mac or Notepad on Windows). Save this file under
nabseq_nf/references/reference_sequences/with a .fasta extension in the format of ORGANISM_whatever.fasta (e.g. rat_all_imgt_refs.fasta)Done!
Building IgBLAST databases
Download reference files Visit the IMGT/V-QUEST website and scroll down until you see IMGT/GENE-DB reference directory sets. Here, you will need to download the V, (D), J and C genes for each of IGH, IGL and IGK.

Combine reference sequences You need to combine all of the V, (D), J and C genes into 4 files (so you should have a V genes file containing IGHV, IGKV, IGLV and a D genes file containing IGHD, and so on). At this stage, you may also like to concatenate the constant sequences that come from the same gene (IMGT reports the CH1, CH2, CH3 etc of each constant region gene separately) although this is not essential (just makes the files a bit cleaner). you should name these files like: imgt_ORGANISM_V_unclean and imgt_ORGANISM_D_unclean etc This is important because NAb-seq will use this naming convention to look for your databases when running IgBLAST.
Clean up the reference sequences Before building the database, you will need to download IgBLAST and run
edit_imgt_file.pl(located in the bin directory) on each of the above files like so:ncbi-igblast-1.19.0/bin/edit_imgt_file.pl imgt_ORGANISM_V_unclean > imgt_ORGANISM_VCreate the databases Finally, you need to run
makeblastdblike so:ncbi-igblast-1.19.0/bin/makeblastdb -parse_seqids -dbtype nucl -in imgt_ORGANISM_VIf the databases don’t work, you can try checking them using theblastdbcheckutility from the BLAST package. One error I came across was missing taxonomy information, which was solved by following the instructions at the bottom of this page. important make sure that these databases are located innabseq_nf/references/igblast/databases/
IgBLAST internal & optional data
If you are using one of the organisms supported by IgBLAST (human, mouse, rat, rabbit and rhesus_monkey), you can get these files from within the standalone IgBLAST program. Guide for setting up custom organisms coming soon!